This web page was created programmatically, to learn the article in its authentic location you possibly can go to the hyperlink bellow:
https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-12088-6
and if you wish to take away this text from our web site please contact us
Multi-sample SV characterization
SVTopo was used to visualise SVs in a set of seven unrelated HiFi genomes: Genome in a Bottle Ashkenazi Trio Son HG002 [27] and 6 publicly launched genomes from the three-generational Platinum Pedigree cohort [28]. These six genomes (NA12889, NA12890, NA12891, NA12892, 20,080, and 20,100) are the unrelated parental genomes in that cohort, thus representing all included ancestral haplotypes within the Platinum Pedigree. All genomes have been aligned to the GRCh38 human reference and phased with WhatsHap v1.4. SV calling was carried out with Sawfish v0.12.7. Sequencing protection was roughly 30 × for HG002 and 200,080, 45 × for 200,100 and NA12890, and 60 × for NA12889 and NA12892. SVTopo generated SV pictures for all samples on a single processor in 8 h, 39 min, about 1.25 h per genome. The most RAM utilization was 4.75 GB.
Excluding easy SV networks (omitted by default filtering on the picture technology step, see Fig. 1), this evaluation resulted in 446 pictures. 125 pictures have been generated with incompletely resolved construction, a lot of that are indicative of alignment artifacts and false-positive SV calls (see Supplementary Figs. 1–5). 92 pictures represented canonical inversions, whereas 37 represented easy pairs of deletion or duplication variants related by phasing. The remaining 192 pictures represented high-confidence complicated SVs. Many of those rearrangements have been shared by a number of samples, with a complete of 101 distinctive complicated SV loci [29]. While well-defined classes don’t exist for many complicated SVs, these variants have been manually assigned to 10 complicated SV teams (see Fig. 3 and Supplementary Table 1), which we created for this variant set to simplify evaluation. Five singleton SVs have been individually distinctive and have been grouped collectively as Unique-singleton variants, with the opposite 9 teams containing structurally related SVs.
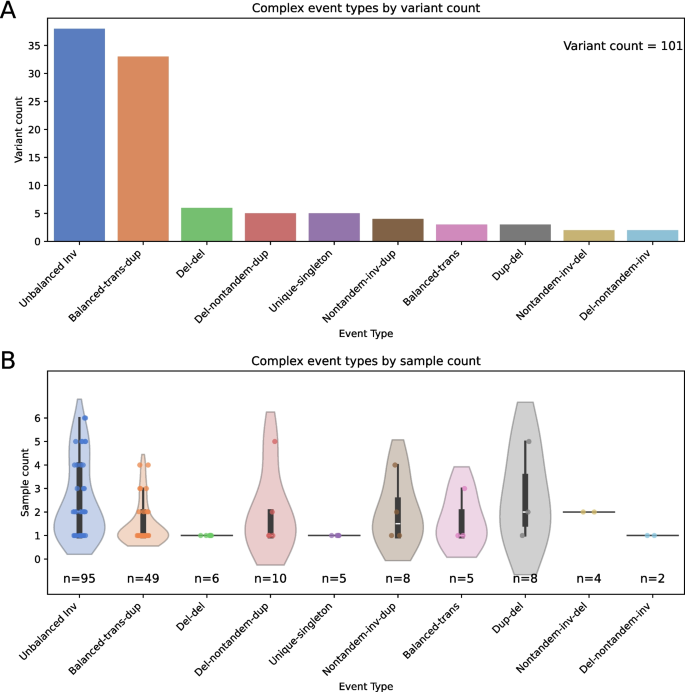
Types and distributions of complicated SVs in seven unrelated genomes: 101 distinctive complicated SVs have been manually assigned to 10 classes comparable to variations on easy SV varieties, with 5 complicated SVs grouped as Unique-singleton. A Counts of distinctive SVs assigned to every class. B Distributions of pattern counts for variants belonging to every SV class
Many of those SVs are difficult to visualise with present instruments however render clearly with SVTopo. This is demonstrated by examples of non-tandem duplications and balanced translocations with flanking duplication, as proven in Fig. 4. See additionally Supplementary Figs. 6–12 for extra examples of those complicated SV varieties with comparability to IGV and Ribbon visualizations, every containing notes to explain the class definition.
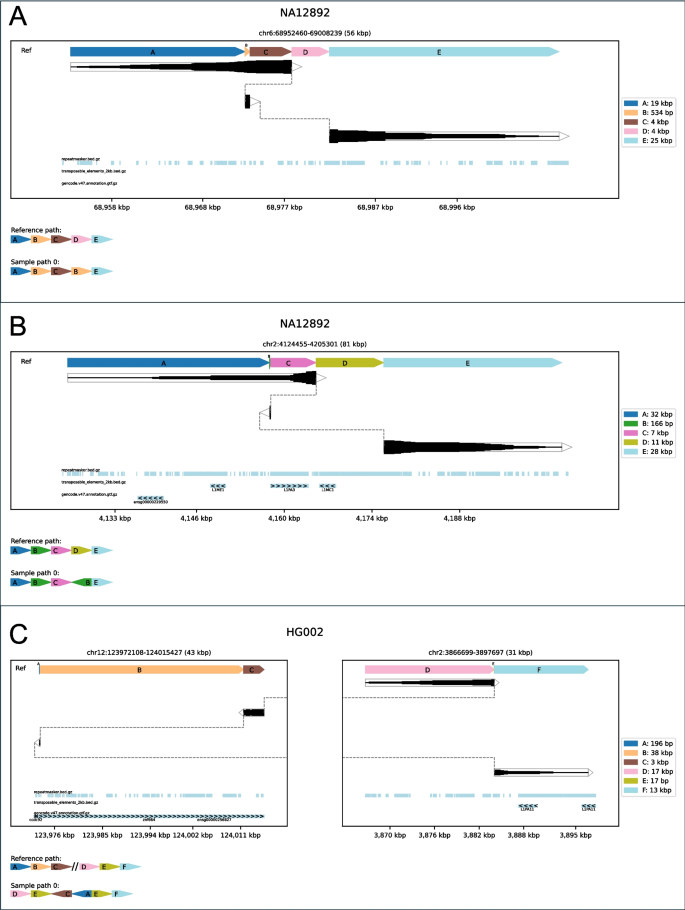
Examples of three complicated SV classes: Plots of three complicated SV varieties which can be difficult to interpret with out SVTopo representations. A Dup-del: non-tandem duplication adopted by a deletion. B Nontandem-inv-del: An inverted non-tandem duplication adopted by a deletion. C Balanced-trans-dup: A balanced translocation consisting of two blocks from chr12 translocated to chr2 and the duplication of a brief area on chr2
Inversions have been the most typical SV sort plotted by SVTopo from these seven genomes (easy deletions and duplications being omitted by default) and have been grouped into three classes: balanced (canonical) inversions (Inv, n = 44), with a genomic block reversed in orientation and straight reconnected; inversions with precisely one flanking deletion (Inv-del, n = 17); and inversions with flanking deletions on each side (Inv-double-del, n = 25). While usually not thought of a posh SV (and omitted from complicated SV classes in Fig. 2), balanced inversions are sometimes fairly difficult to visualise utilizing commonplace methods. Visualizations not designed for complicated SVs wrestle to indicate every of the part rearrangements in inversions distinctly, notably for unbalanced inversions (See Fig. 5).
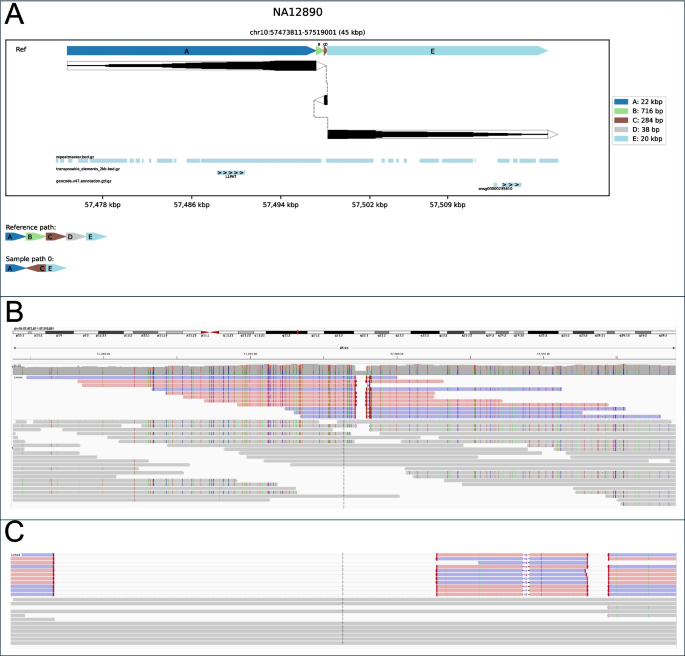
Example unbalanced inversion with flanking deletions: A 284 bp inversion flanked by a 716 bp deletion and a 38 bp deletion (class Del-inv-del). A SVTopo plot of area. B IGV display screen seize of the area with linked supplementary alignments. C IGV picture of the area, zoomed in to incorporate solely complicated SV breakpoints
A complete of 44 distinctive balanced inversion variants have been plotted, 19 occurring in a number of samples to look a complete of 83 occasions. 17 distinct inversions with a single flanking deletion have been plotted, showing a complete of 42 occasions; and 25 with two flanking deletions, showing 52 occasions. Almost half (42 of 86) of the inversions have been accompanied by flanking deletions (see Fig. 6 and Supplementary Table 2).
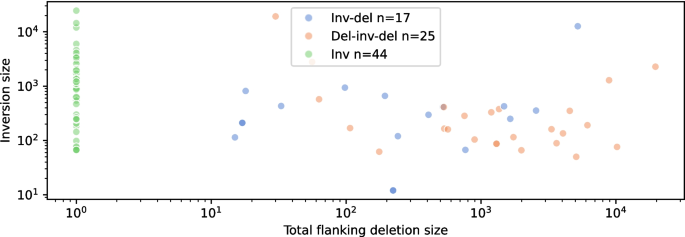
Balanced and unbalanced inversion sizes: Lengths of inverted blocks contrasted towards flanking deletion sizes for inversions with 0–2 flanking deletions. Each axis is proven on log scale. Non-tandem inversions excluded
SVTopo desk viewer
In addition to offering easy visualizations for SV evaluate, SVTopo routinely creates a feature-rich serverless HTML-based viewing desk to prepare and current pictures (see Fig. 7). This viewer is a self-contained internet web page, with built-in filtering instruments to permit customers to quickly establish the complicated SVs most related to a analysis precedence (similar to reviewing pictures related to a area or pattern of curiosity). The viewer might be deployed regionally for a single person or remotely if multi-user entry is desired (instance viewer publicly out there [30]). The viewer foremost web page consists of a variant area desk, with a row for every genomic window represented. Clicking a row shows the picture. Columns within the desk present the chromosome, begin, finish, area measurement, related variant IDs (if out there), variety of variant IDs represented within the picture (if out there), and pattern ID. The desk permits a person to filter by clicking a variant ID, variety of variants discipline, or pattern ID. A filtering choices window additionally permits filtering on area measurement ranges and goal chromosomes.
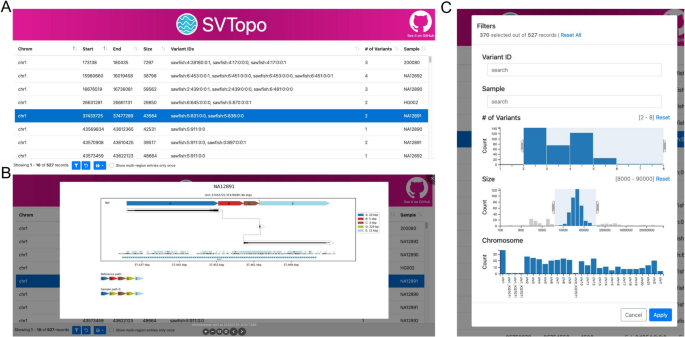
SVTopo desk viewer. A A display screen seize of the desk viewer, exhibiting included desk columns. B A display screen seize of a posh SV picture displayed by clicking a desk row. Image reveals a deletion and non-tandem duplication. C Viewer filtering window, with filtering fields for variant ID, pattern ID, variety of variants, area measurement, and chromosome. Filters proven choose for two–8 variants per picture, a measurement vary of 8,000–90,000 bp, and all chromosomes
This web page was created programmatically, to learn the article in its authentic location you possibly can go to the hyperlink bellow:
https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-12088-6
and if you wish to take away this text from our web site please contact us


