This web page was created programmatically, to learn the article in its authentic location you may go to the hyperlink bellow:
https://www.nature.com/articles/s41467-025-64750-y
and if you wish to take away this text from our web site please contact us
Parasitaemias in cattle comprise combined populations expressing each slender- and stumpy-associated transcriptomes
Two calves had been contaminated by way of the intravenous route with 1 × 106 T. b. brucei Antat 1.1 90:13, and blood parasitaemia was monitored over 60 days (Fig. 1A, Supplementary Fig. 1). Within the decision of research, parasitaemia peaked between days 5 and 23 put up an infection, reaching an noticed most of 4 × 104/ml and three × 105/ml in cows 1 and a couple of, respectively, earlier than coming into a power part characterised by parasitaemia <1 × 104/ml after day 23. In this power part, for each cattle, parasitaemia continued principally between 1–3 × 103/ml, or sometimes fell beneath the brink of detection. These parasitaemia dynamics and variability between calves are in line with earlier experimental infections of cattle with the identical parasite pressure20.
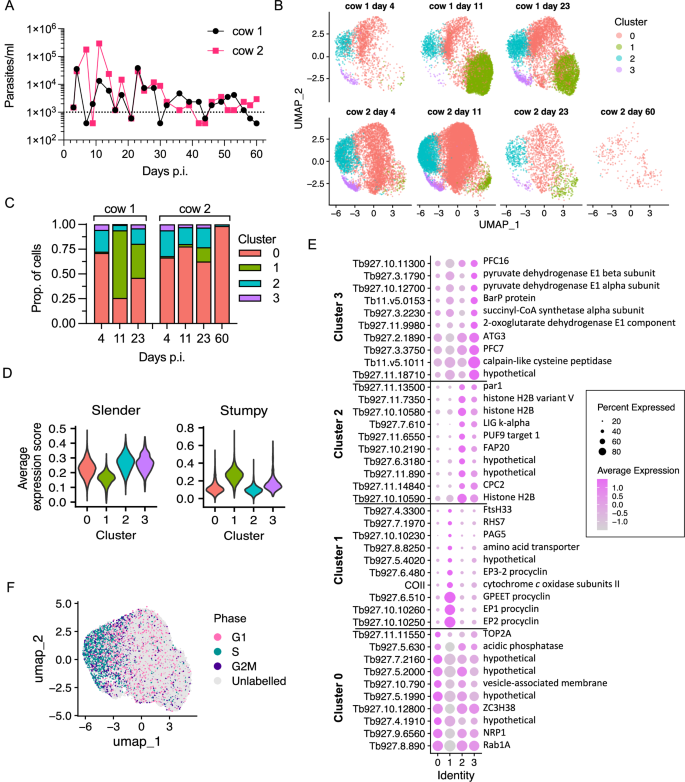
A Blood parasitaemia ranges for cow 1 (black) and cow 2 (pink). Dotted line signifies the low confidence threshold. B Clustered particular person transcriptomes of T. brucei remoted on days 4, 11, 23 and 60 (solely cow 2) put up an infection. Cluster colors are constant throughout plots. C Proportion of every pattern, cut up by cow, in every of the clusters. D Average expression scores of slender (left) and stumpy (proper) marker genes for every cluster. Scores above 0 point out increased common expression of the marker gene set recognized beforehand21, in comparison with a set of randomly choose management genes. Cluster colors as in (B). E Dot plot of the typical expression of the highest ten marker genes for every cluster with the very best fold change (color scale). The dimension of every dot is proportional to the share of cells in that cluster expressing the marker. Gene title or description is proven to the best. F UMAP of all samples colored by cell cycle part. “Unlabelled” signifies the cell didn’t overexpress any part marker genes (G1, S or G2/M), in comparison with a random set of management genes. Source information are supplied as a Source information file.
To examine the doubtless heterogeneous populations of T. brucei within the cattle bloodstream, we used scRNA-seq at time factors when adequate parasites could possibly be remoted and purified; days 4, 11 and 23. Parasites had been additionally purified from the blood on the finish of the experiment (day 60) though extraordinarily low parasite numbers at the moment level hampered efforts to acquire adequate cells for Chromium scRNA-seq evaluation. Despite this, we obtained between 1585 and 13,747 particular person transcriptomes from T. brucei for each cows on days 4, 11, and 23, in addition to 257 transcriptomes from cow 2 on day 60 (Supplementary Fig. 2). Transcriptomes obtained from cow 1 on day 60 didn’t meet high quality management thresholds for evaluation and had been excluded.
Samples had been built-in collectively earlier than dimension discount and clustering evaluation (Fig. 1B). Four distinct clusters had been recognized, and these had been detected in all samples excluding cow 2 day 60 (Fig. 1C) the place the low variety of transcriptomes captured restricted evaluation. Independent approaches recognized that three of the clusters (0, 2 and three) had a transcriptome in line with slender varieties however assorted in different elements, whereas cluster 1 (inexperienced) contained transcriptomes that extra carefully resembled stumpy varieties. Firstly, the typical expression stage of beforehand recognized slender-associated markers21 was decrease in cluster 1 in comparison with the opposite clusters and, conversely, the typical expression of stumpy-associated markers was increased in cluster 1 in comparison with clusters 0, 2 and three (Fig. 1D). Secondly, de novo identification of marker genes related to every cluster was performed by way of differential expression evaluation between the clusters (Supplementary information 1, Supplementary Fig. 3). The ten markers with the very best fold change had been plotted for every cluster (Fig. 1E). This included procyclin encoding transcripts, the highest markers of cluster 1. This was adopted by gene ontology (GO) time period enrichment to determine organic processes related to every cluster (Supplementary Fig. 3B, Supplementary information 1), mentioned intimately beneath. Thirdly, the cell cycle part of every cell was categorised utilizing the expression of beforehand outlined part marker genes22 (Fig. 1F). The common expression of G1, S and G2/M marker genes was calculated per cell and in comparison with the typical expression of a set of random management genes to generate an “average expression score” for every part (Supplementary Fig. 4A). Cells had been labelled with the part with the very best expression rating, or “Unlabelled” if common expression was not above that of the management gene set for any part.
Slender-like clusters 2 and 0 had been largely distinguished by variations of their cell cycle part proportions (Fig. 1F, Supplementary Fig. 4B). Cluster 2 confirmed excessive proportions of S (46%) and G2/M (13%) part labelled cells, in line with the highest marker genes (i.e. chromosome passenger advanced 2 and histones, Fig. 1E) and related GO phrases that embrace “DNA packing” and “cytokinesis” (Supplementary Fig. 3B). Hence, cluster 2 consisted of proliferative slender varieties. Cluster 0 in distinction was largely made up of “Unlabelled” cells (79%) and had the fewest distinctive markers (Supplementary Fig. 3A), with the highest markers additionally detected within the different slender-like clusters 2 and three (Fig. 1E). GO phrases for cluster 0 embrace “purine nucleoside disphosphate metabolic process” and “phosphorylation” on account of expression of slender-related genes together with hexokinase and pyruvate kinase 1 (Supplementary Fig. 3B, Supplementary information 1). This inhabitants therefore seems to include few parasites in S or G2/M phases, however retains expression of different slender-associated markers.
Cluster 3 contained cells in all cell cycle phases, with 17% and 15% residing in S and G2/M, respectively, however with a big proportion of unlabelled cells, indicating it was made up of each dividing and non-dividing parasites (Supplementary Fig. 4B). This cluster was distinguished by a definite set of marker genes referring to “tricarboxylic acid cycle” and “pyruvate metabolic process” (Fig. 2A), together with mitochondrial malate dehydrogenase and 2-oxoglutarate dehydrogenase (Fig. 2B). This subpopulation of slender-like varieties had upregulated the expression of genes required to additional metabolise pyruvate to generate ATP (Fig. 2B, C), distinct from the classically described secretion of pyruvate by slender varieties23, however maintained and in some instances upregulated expression of glycolytic elements (Fig. 2C).
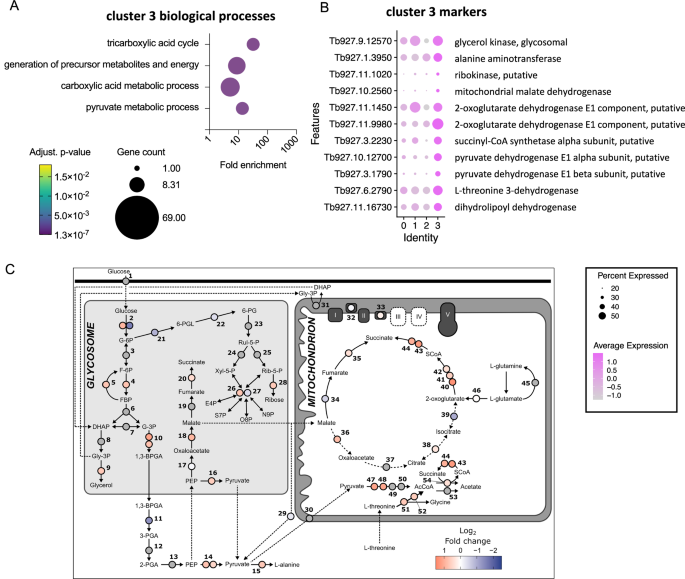
A Biological course of GO phrases enriched for cluster 3 marker genes. Plot indicated the fold enrichment (x-axis), variety of genes for every time period (dimension) and the adjusted p-value (color scheme) for every time period (Benjamini-Hochberg adjusted p-values from Fisher’s precise check). B Dot plot of the typical expression of chosen cluster 3 markers genes linked to phrases “pyruvate metabolic process” and the “TCA cycle”. C Log2 fold change (Cluster 3 in comparison with clusters 0, 1 and a couple of mixed) was calculated for every gene. Dashed traces signify transport processes. Grey circles point out genes weren’t considerably completely different in cluster 3 relative to different clusters. Genes: 1, hexose transporters; 2, hexokinase, Tb927.10.2010 and Tb11.v5.0732; 3, glucose-6-phosphate isomerase; 4, phosphofructokinase, Tb927.3.3270; 5, fructose-1,6-bisphosphatase, Tb927.9.8720; 6, aldolase; 7, triosephosphate isomerase; 8, glycerol-3-phosphate dehydrogenase; 9, glycerol kinase, Tb927.9.12570; 10, glyceraldehyde 3-phosphate dehydrogenase, Tb927.6.4280 and Tb927.6.4300; 11, phosphoglycerate kinase, Tb927.1.710; 12, phosphoglycerate mutase; 13, enolase; 14, pyruvate kinase 1, Tb927.10.14140 and Tb11.v5.0605; 15, alanine aminotransferase, Tb927.1.3950; 16, pyruvate phosphate dikinase, Tb927.11.6280; 17, Phosphoenolpyruvate carboxykinase, Tb927.2.4210; 18, glycosomal malate dehydrogenase, Tb927.10.15410; 19, glycosomal fumarate hydratase; 20, glycosomal NADH-dependent fumarate reductase, Tb927.5.930; 21, glucose-6-phosphate dehydrogenase, Tb927.10.2490; 22, 6-phosphogluconolactonase, Tb927.11.6330; 23, 6-phosphogluconate dehydrogenase; 24, ribulose-5-phosphate epimerase; 25, ribose 5-phosphate isomerase; 26, transketolase, Tb927.8.6170; 27, transaldolase, Tb927.8.5600; 28, ribokinase, Tb927.11.1020; 29, malic enzyme, Tb927.11.5450; 30, Mitochondrial pyruvate provider 2; 31, FAD-dependent glycerol-3-phosphate dehydrogenase; 32, NADH dehydrogenase (NDH2), Tb927.10.9440; 33, Alternative oxidase, Tb927.10.7090; 34, mitochondrial fumarate hydratase, Tb927.11.5050; 35, mitochondrial NADH-dependent fumarate reductase, Tb927.10.3650; 36, mitochondrial malate dehydrogenase, Tb927.10.2560; 37, citrate synthase; 38, aconitase,Tb927.10.14000; 39, isocitrate dehydrogenase, Tb927.8.3690; 40, 2-oxoglutarate dehydrogenase E1 part, Tb927.11.9980; 41, 2-oxoglutarate dehydrogenase E1 part, Tb927.11.1450; 42, 2-oxoglutarate dehydrogenase E2 part, Tb927.11.11680; 43, succinyl-CoA synthetase α, Tb927.3.2230; 44, succinyl-CoA ligase β, Tb927.3.2230; 45, glutamine synthetase; 46, glutamate dehydrogenase, Tb927.9.5900; 47, pyruvate dehydrogenase E1 α subunit, Tb927.10.12700; 48, pyruvate dehydrogenase E1 β subunit, Tb927.3.1790; 49, dihydrolipoamide acetyltransferase; 50, pyruvate dehydrogenase advanced E3; 51, L-threonine 3-dehydrogenase, Tb927.6.2790; 52, 2-amino-3-ketobutyrate coenzyme A ligase, Tb927.8.6060; 53, Acetyl-CoA hydrolase (ACH), Tb927.3.4260; 54, Succinyl-CoA:3-ketoacid coenzyme A transferase (ASCT), Tb927.8.6060. Source information are supplied as a Source information file.
In distinction, cluster 1 was related to “translation” (Fig. 3A) as a result of excessive variety of ribosomal protein transcripts that confirmed elevated ranges on this cluster, in addition to eukaryotic translation initiation issue 5 A (EIF5A) and receptor for activated C kinase 1 (RACK1)24. Notably, earlier transcriptomic evaluation of T. b. rhodesiense remoted at peak parasitaemia in rats, when stumpy varieties are dominant, additionally confirmed excessive ranges of translation related transcripts25. Cluster 1 markers (Fig. 3B, Supplementary information 1) additionally included: Protein Associated with Differentiation (PAD) members of the family which might be concerned within the notion of the stumpy-to-procyclic differentiation sign26; procyclic type floor proteins EP and GPEET procyclin27; genes encoded on the maxi circle kDNA, together with a mitochondrial NADH dehydrogenase subunit ND4 and cytochrome oxidase subunit II; the stumpy-elevated and warmth stress related RNA regulator ZC3H1128; the iron responsive regulator RBP5 (Tb927.11.12100)29; Factor H receptor30; and, the pteridine transporter Tb927.10.9080, which is a goal of snoGRUMPY, a proposed regulator of stumpy formation31. Therefore, cluster 1 parasites expressed identified transcript markers of stumpy varieties, and had been strongly related to elevated expression of genes required for international translation and developmental gene regulation. This subpopulation is henceforth known as stumpy-like for these causes.
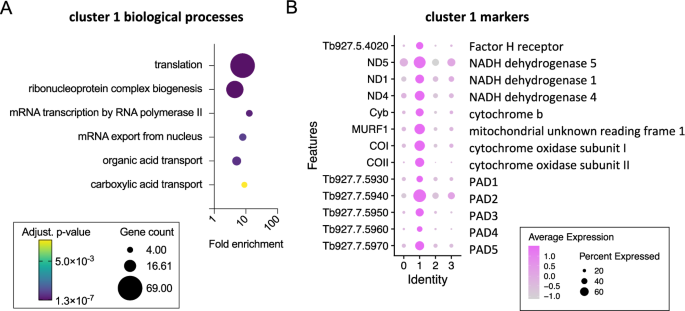
A Biological course of GO phrases enriches for cluster 1 marker genes. Plots point out fold enrichment (x-axis), variety of genes for every time period (dimension) and the adjusted p-value (color scheme) for every time period (Benjamini-Hochberg adjusted p-values from Fisher’s precise check). B Dot plot of the typical expression of cluster 1 marker genes beforehand linked to stumpy varieties.
The look of parasites expressing stumpy-associated transcripts correlates with diminished proliferation and shorter flagella lengths
When attainable, parasites had been additionally examined by microscopy. Given the looks of a subpopulation expressing stumpy-associated transcripts and the low variety of parasites out there, we prioritised analysing hallmarks of stumpy type growth: cell division cycle modifications, PAD1 floor protein expression, and altered morphology by way of diminished flagellum size (Fig. 4).
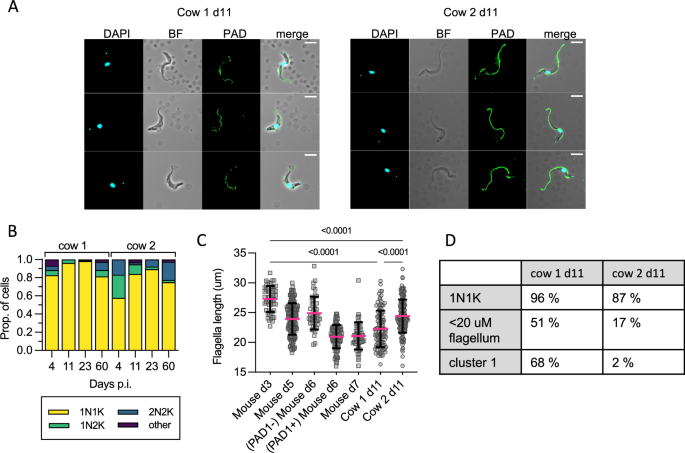
A Representative cell photos of parasites remoted on d11 p.i. in cow 1 and cow 2, displaying an absence of cell floor PAD1 staining (besides the flagella). B KN counts of parasites in every cow at every time level. C The distribution of flagellar measurements from parasites in each cows on d11 p.i. in comparison with controls taken from mice. On d3 (n = 1, on d3 p.i. blood parasites are absent in most mice), all mouse parasites are slender, PAD1 damaging, and totally replicative; on d5 (n = 3) all seem slender, are PAD1 damaging, and just some are replicative, on d6 (n = 3) some parasites are PAD1 constructive, only a few are replicative; on d7 (n = 3) all are PAD1 constructive and non-replicative. Parasites from the cows on d11 had been considerably completely different from d3 true slender varieties from mice, and from one another. P-values indicated present the results of two-sided Dunn’s a number of comparability checks between samples. Data are offered as imply +/− SD. D A comparability desk of the proportion of cells belonging to stumpy-like cluster 1, the proportion having a flagellar size most much like mouse derived stumpy varieties (< 22 μm) and variety of non-dividing (1K1N) cells, displaying the distinction between cow 1 and cow 2 on d11 p.i. Source information are supplied as a Source information file.
Dividing parasites first segregate the mitochondrial kDNA (Ok) adopted by the nuclear genome (N) permitting cell cycle standing to be inferred from the KN configuration. Parasites with segregated kDNA and 1 nucleus, 2K1N, signify these in late S part or G2, whereas parasites with segregated kDNA and a couple of nuclei, 2K2N, are post-mitotic. Proliferative parasites with 1 kinetoplast and 1 nucleus (1K1N) could be in G1 or early S part, whereas stumpy cells have exited the cell cycle and uniformly exhibit 1K1N. The parasites remoted from cow 2 on day 4, because the parasitaemia was quickly ascending in the direction of its peak within the first wave of an infection, had been enriched for dividing varieties (43% 2K1N or 2K2N, Fig. 4B). All different samples profiled contained 2K1N/2K2N parasites, albeit in low numbers, indicating that the cattle blood contained dividing T. brucei at every level however as a really small proportion of the general inhabitants relative to the institution part. Notably, the proportion of 1K1N parasites reached 96% and 98% in cow 1 on days 11 and 23, respectively, indicating low ranges of division in line with stumpy growth13. These samples additionally contained the best proportions of cluster 1 parasites recognized by scRNAseq evaluation (Fig. 1C).
Further makes an attempt to quantify the proportion of proliferative parasites had been made utilizing an ex vivo plating assay, which beforehand allowed correct detection of replication-competent parasites when current in as few as 0.1% of the entire inhabitants13. Parasites had been seeded into 96-well plates at identified dilutions (50, 20, 5, 2 and 1 parasite(s)/nicely), with twelve wells ready per dilution. Surprisingly, no samples gave rise to proliferating cultures excluding parasites remoted from cow 2 on day 4, by which 5/12 wells grew at 20 parasites/nicely seeding (estimated 3% viability primarily based on chance equation, see strategies), 4/12 wells at 5 parasites/nicely (11% viability) and a couple of/12 wells at 2 parasites/nicely (9% viability). Thus, it was estimated solely 7.7% of parasites remoted from cow 2 on day 4 had been viable and in a position to replicate ex vivo regardless of the massive proportion of actively dividing cells (as outlined by their 2K1N or 2K2N standing). Given the detection of lively cell division markers by each microscopy and scRNA-seq in all cattle samples, we think about that speedy adaptation to the bovine setting rendered the parasites unable to proliferate in tradition media as soon as remoted, no matter their replicative capability in vivo.
As the proportion of stumpy-like cluster 1 parasites assorted most between the 2 cows on day 11 (Fig. 1C), we used this time level to correlate two established differentiation parameters with the looks of this cluster: stumpy-specific protein PAD1 expression26 and flagellar shortening that marks each intermediate and stumpy levels32. No parasites confirmed expression of PAD1 throughout the parasite floor in both pattern. Note that PAD1 antiserum can bind the flagella of parasites no matter their differentiation standing, and this contains monomorphic pressure Lister 427 (Supplementary Figs. 5 and 6). In mouse infections, flagellar size supplied a sign of morphological differentiation, being shorter in stumpy varieties, however there was additionally a quantifiable change between slender varieties from proliferating populations (imply = 27 µm) and differentiating populations (imply = 24 µm) that had been but to tackle a full stumpy morphology (imply = 19 µm)32. Visual inspection of dwell parasites from cattle revealed that the majority parasites had a slender to intermediate morphology (see Supplementary video 1). Flagella size was used to quantify this morphological change and this was in comparison with the identical parasite stabilate present process slender-to-stumpy morphological growth in mice, thereby calibrating this transition in a bovine an infection with occasions occurring within the nicely characterised murine mannequin (Fig. 4C). The shortening of the flagellum was evident throughout mouse an infection, as anticipated, because the parasites transitioned from a proliferative slender inhabitants (day 3, imply size 27.3 µm) to a differentiating inhabitants of non-proliferating varieties that didn’t but resemble full stumpy varieties (day 5, imply size 23.9 µm), a heterogeneous inhabitants of PAD1 damaging differentiating varieties (day 6, imply 24.9 µm) and PAD1 constructive stumpy varieties (day 6, imply 21 µm), earlier than all cells had been PAD1 constructive stumpy cells on d7 (imply = 21 µm). Notably, most parasites remoted from each cattle on day 11 had a diminished flagellar size (means = 22.3 µm and 24.4 µm, respectively), extra in line with differentiating varieties than proliferating slender varieties. Additionally, the flagellum size distribution differed between cow 1 and cow 2 on day 11, with extra parasites with shorter flagella in cow 1, in line with a extra differentiated inhabitants. In distinction in cow 2, with extra proliferative parasites, flagella had been longer. This correlates with the upper proportion of cells belonging to stumpy-like cluster C1 and with a 1K1N configuration in cow 1 than in cow 2 (Fig. 4D).
Thus, the brief flagellum, prevalence of cluster 1 cells, and people with a 1K1N profile, every are indicative of a prevalence of stumpy-like parasites in cattle blood that, a minimum of within the samples investigated, didn’t categorical PAD1 protein.
In power infections, parasite morphology revealed combined populations of each dividing and differentiating varieties
Having established the cytological traits of the stumpy-like varieties (primarily based on their transcriptome), we used these options to research the bloodstream varieties current in power cattle an infection when parasitaemia ranges dropped to round 1 × 103–1 × 104 parasites/ml, beneath the extent possible for efficient scRNA-seq evaluation.
In this power stage, parasites had been overwhelmingly 1K1N, notably between days 49–58 (imply = 93.4% 1K1N, min = 89.4, max = 97.6; Fig. 5A, B). This was additionally noticed when the parasitaemia was extraordinarily low, indicating low ranges of lively parasite division within the blood throughout many of the power part of an infection. Despite the vast majority of parasites being non-dividing varieties, solely 2/824 parasites had been noticed that stained positively for PAD1, each from cow 2 on d58, which was in line with the overall absence of totally morphologically stumpy varieties within the power part parasite inhabitants (Fig. 5A).
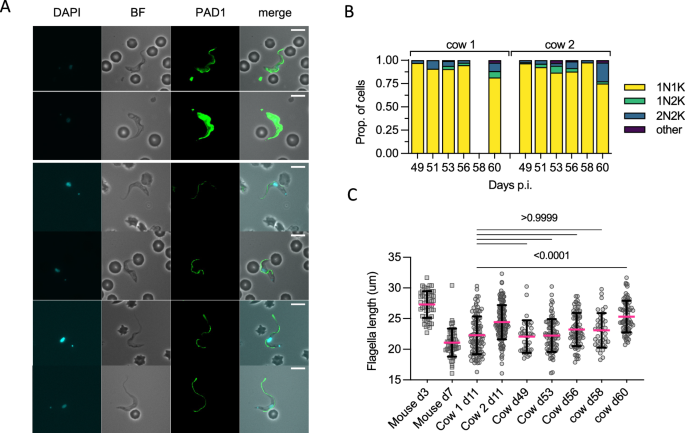
A Representative cell photos of parasites remoted from d49 to d60 p.i. Only two parasites (each proven) had been scored as PAD1 constructive, with most cells displaying comparable slender-intermediate morphology of these seen on d11 p.i. B Kinetoplast (Ok) and nucleus (N) configuration counts for every pattern. C The distribution of flagella measurements in each cows within the power part vs controls from d11 p.i. and mouse slender varieties (day 3, n = 1 mouse) or stumpy varieties (day 7, n = 3 mice). P-values indicated present the results of two-sided Dunn’s a number of comparisons checks between cow d11 p.i. (which comprises a excessive proportion of stumpy-like varieties) and the samples taken within the late stage of an infection (mixed cow 1 and cow 2 replicates). Data are offered as imply +/− SD. Source information are supplied as a Source information file.
To look at their developmental standing additional, the flagella size of power part parasites was in comparison with that of cow 1 day 11, when a excessive proportion of stumpy-like parasites was evident by single cell RNA evaluation (Fig. 5C). No important distinction in flagella size was discovered between these time factors, excluding day 60 when the imply flagella size was longer (imply for each cows = 25.33 µm). Consistently, this time level was characterised by having the next proportion of 2K2N cells in each cows. Taken as a gaggle, nonetheless, the flagellar size of power parasites was most much like combined populations of parasites containing each actively dividing and non-dividing differentiating parasites, much like the cluster 1 stumpy-like inhabitants.
Thus, the power part of infections contained massive proportions of parasites with the shorter flagella related to stumpy-like varieties albeit with out PAD1 protein expression, with a minority of actively dividing varieties and scarce PAD1-positive morphologically stumpy varieties.
Host particular distinction in T. brucei transcriptomes
To discover host-specific variations of the parasite populations, we in contrast the transcriptomes of parasites in every of the cow samples to our earlier single-cell transcriptomic information generated from both in vitro culture-derived21 or mouse-derived parasites13 by pseudobulking, whereby transcript counts for every gene had been summed throughout each cell within the inhabitants inside that pattern. The ‘In vitro’ samples had been combined populations of slender and stumpy varieties generated by publicity to mind coronary heart infusion broth (BHI) to set off stumpy growth21. Mouse derived samples contained a mixture of slender, differentiating and, most prominently, stumpy type parasites remoted on days 7 and 23 of an infection13.
For preliminary principal part evaluation (Fig. 6A), variant floor glycoproteins (VSGs) had been faraway from the genes used to calculate principal elements to keep away from grouping by VSG expression, which is predicted to fluctuate throughout an infection. Differential expression checks had been then carried out with DESeq2 between the completely different host environments (Fig. 6B–D). All samples had been used for every situation (in vitro, mouse or cow) in these comparisons no matter time level or an infection part.
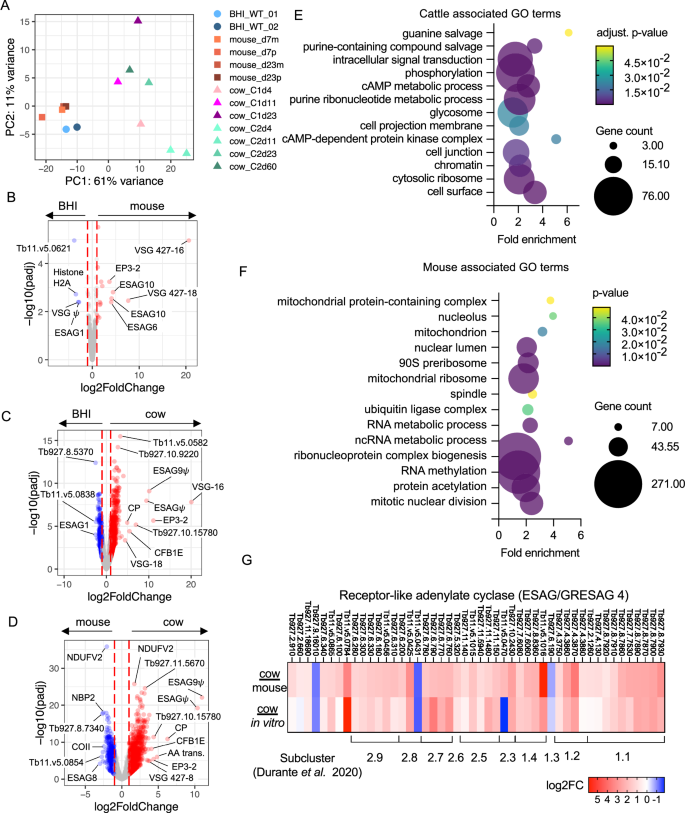
A PCA plot of all in vitro (circles, ref. 21), mouse (squares, ref. 13) and cattle (triangles) derived single cell transcriptomic samples, which were pseudo bulked. Volcano plots of adjusted p worth (y-axis, −log10(padj) and fold change (x-axis, log2(fold change)) of differentially expressed genes between pseudo-bulked (B) BHI handled in vitro and mouse derived samples, C BHI handled in vitro and cattle derived samples and D mouse and cattle derived samples. In every case all samples and time factors, as proven in (A), are used for every host setting and padj values are Benjamini-Hochberg adjusted p-values from Wald checks. E Enriched GO phrases in genes over expressed in cattle derived samples in comparison with mouse derived samples. F Enriched GO phrases in genes over expressed in mouse derived samples in comparison with cattle derived samples. GO time period adjusted p values are Benjamini-Hochberg adjusted p-values from Fisher’s precise check. G Fold change (log2) in ESAG4/GRESAG4 transcript ranges encoding receptor-like adenylate cyclase proteins, for parasites remoted from cattle relative to mice (prime) in vitro tradition (backside). Clade and subcluster of every gene beforehand recognized33 are proven beneath.
From the PCA evaluation, it was obvious that T. brucei from mouse and tradition techniques had been similar to one another, though 55 genes had increased expression and 32 confirmed decrease expression in mouse samples when in comparison with parasites uncovered to the BHI quorum-sensing sign in vitro (Fig. 6B, Supplementary information 2). Of these, 9 differentially expressed genes encoded completely different expression web site affiliate genes (ESAGs) and VSGs, maybe reflecting use of an alternate expression web site. ESAG10, which encodes folate transporters, and ESAG6, transferrin receptor subunit, had been overexpressed in mouse derived samples, whereas ESAG1 was downregulated in comparison with BHI generated samples.
In distinction, the transcriptome of cattle derived parasites was clearly distinguished from these of mice and tradition (Fig. 6A). Most variance was defined by PC1 (61%), which separated cow samples from mouse or tradition samples. Additionally, there was variation between samples taken over the course of cattle an infection, and between particular person cows on the similar time level. Strikingly, after 23 days of mouse an infection the transcriptome of cells remained carefully associated to cultured parasites, even when the proportions of slender and stumpy varieties assorted between samples. In distinction, parasites from cattle could possibly be discriminated from mice and tradition derived parasites primarily based on their transcriptome as quickly as 4 days after an infection, demonstrating speedy gene expression modifications, almost certainly associated to host adaptation, following bovine an infection.
Cattle derived samples had far higher numbers of differentially expressed genes compared to each in vitro (Fig. 6C) and mouse derived (Fig. 6D) samples. GO time period enrichment for genes with increased transcript ranges in cattle derived samples (1678 adjusted p < 0.05 and fold change >2), confirmed enrichment for transcripts related to elements of the cell floor, cytosolic ribosomes, chromatin, the flagellum and the glycosome (Fig. 6E). GO phrases related to genes that confirmed increased transcript ranges in mouse derived pattern largely associated to ribonucleoprotein advanced biogenesis, as nicely at nuclear division and the mitochondrion (Fig. 6F). Predicted cell floor proteins that assorted of their expression between infections of mouse or cattle hosts included expression web site related genes (ESAGs). The ESAG courses that assorted mostly between host species had been ESAG10 (3/7 annotated ESAG10 genes with detectable transcripts differentially expressed between mouse and cow), ESAG4 (7/18 genes), ESAG2 (9/25 genes) and ESAG8 (3/11 genes) (Supplementary information 2). The ESAGs with the best fold-change in transcript ranges had been two pseudogenes upregulated in cattle samples, ESAG9 (Tb927.11.110) and a non-classified ESAG (Tb929.11.130), which might be positioned collectively in a probable VSG expression web site (Fig. 6D). The ESAGs with highest expression in mouse samples compared to cattle are additionally positioned in the identical VSG expression web site, Tb427.BES112 (Fig. 6D and Supplementary information 2). Strikingly, GRESAG4 (gene associated to ESAG4), that are positioned within the core chromosomes and never the VSG expression websites, had been additionally upregulated in cattle derived parasites (Fig. 6G). The ESAG4 and GRESAG4 gene household encodes receptor-like adenylate cyclase proteins (ACs), which could be phylogenetically clustered into two clades and additional subgroups33. Clades 1 and a couple of include ACs discovered to be translationally upregulated in bloodstream and procyclic varieties, respectively, by ribosomal profiling34. Transcripts encoding ACs of each clades had been upregulated in cattle derived trypanosomes (31.8% clade 1 and 33.7% clade 2) relative to samples from mouse infections and in vitro tradition. Notably, transcripts encoding ACs termed ACP1-6 which might be identified to localise to the flagellum35 and in some instances have been linked to social motility36 weren’t differentially expressed between host environments. Thus, the noticed modifications in AC transcript expression ranges right here don’t look like linked to life cycle type growth. An various chance is that AC upregulation is linked to T. brucei adaptation or response to the host immune response. In early an infection in mice, the exercise of AC enzymes launched by lysed parasites inhibits the manufacturing of trypanosome-suppressing TNF-α in liver myeloid cells, permitting T. brucei to manage host early innate immune37.
Host particular variations between developmental varieties
Given the clear distinction between hosts, we subsequent sought to grasp developmental form-specific variations between host environments. Attempts to totally combine information from in vitro, mouse and cattle experiments had been inconsistent throughout integration strategies. In the absence of “ground-truth” for these strategies we as a substitute used a label switch method to foretell the cell sorts within the cattle information (question dataset) utilizing both in vitro (Fig. 7A, ref. 21) or mouse (Fig. 7B, ref. 13) parasite cluster labels from our earlier analyses because the reference datasets. Providing proof for the validity of utilizing label switch, a comparability of monomorphic slender type transcriptomes22 to in vitro derived slender and stumpy varieties21 demonstrated that 97.9% of monomorphic slender varieties labelled as slender, as anticipated (Supplementary Fig. 7).
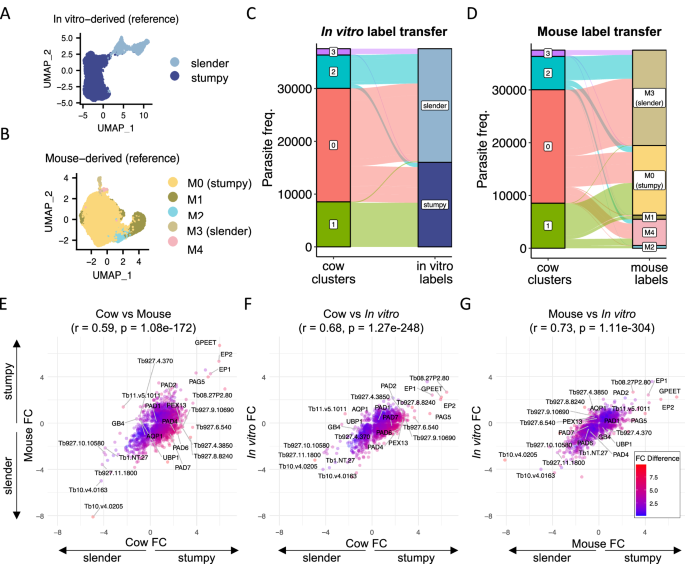
A UMAP of beforehand described scRNA-seq evaluation of T. brucei varieties uncovered to BHI to induce stumpy growth in vitro21. B UMAP of beforehand described scRNA-seq evaluation of T. brucei remoted from the mouse bloodstream used as a reference for label switch13. Slender (M3, gentle brown) and stumpy (M0, yellow) clusters are highlighted. C Proportions of every cattle-derived cluster (left) and the corresponding predicted cell kind utilizing in vitro reference (proper, as proven in A). D Proportions of every cattle-derived cluster (left) and the corresponding predicted cell kind utilizing the mouse reference (proper, as proven in B). Fold change in transcript ranges between stumpy- and slender-like cell sorts for cow vs. mouse (E) cow vs. in vitro (F) and mouse vs in in vitro (G). In every case, outcomes from the Pearson correlation check (R), together with the corresponding two-sided t-test p-value, are proven within the title. Points are colored by the distinction in fold change between the 2 in contrast datasets.
Consistent with our earlier evaluation (Fig. 1D), predicting the cell sorts utilizing the in vitro noticed developmental varieties because the reference (Fig. 7A) resulted within the labelling of cattle cluster 1 as stumpy varieties and clusters 2 and three as slender varieties (Fig. 7C). Cluster 0 was labelled as a mixture of slender and stumpy varieties when in comparison with the in vitro reference, seemingly as a result of low expression of cell cycle marker genes (Fig. 1F). Previous evaluation of T. brucei remoted from the mouse bloodstream on days 7 and 23 of an infection13 revealed that just about all parasites had been arrested stumpy-like varieties at these time factors, with solely 2.1% of the captured parasites being slender varieties (cluster M3, Fig. 7B). In that examine a transparent stumpy inhabitants (cluster M0) was recognized as the commonest type within the mouse bloodstream (82.8%), and two different clusters (clusters M1 and M2) had been recognized which will signify “intermediate” varieties transitioning between slender and stumpy extremes13. Using the label switch technique to foretell the corresponding cell sorts between cattle and mouse derived parasite samples, we recognized 70.9% of cattle cluster 1 (stumpy-like) as most much like mouse cluster M0 stumpy varieties (Fig. 7D). Smaller proportions of cattle cluster 1 had been assigned as cell sorts M1 (7.3%) or M2 (5.3%), suggesting these putatively intermediate parasite varieties had been current however in low proportions in cows on the factors of an infection analysed. Therefore, the stumpy-like varieties discovered within the cattle bloodstream had been most comparable when it comes to their transcriptome to mature stumpy varieties present in mouse infections, regardless of the variations in morphology and lack of PAD1 expression. Matching the comparability to the in vitro information, label switch for cow cluster 0 resulted in a mixture of slender (M3) and stumpy (M0) labels. Additionally, a proportion of all cattle clusters preferentially labelled as mouse cluster M4 (Fig. 7D). Mouse inhabitants M4 was noticed in decrease ranges in earlier murine experiments (Fig. 7B) and organic processes linked to this distinct subpopulation had been unclear13.
The transcript stage fold change between stumpy/stumpy-like (constructive fold change (FC) values) and slender/slender-like (damaging FC values) populations was subsequent quantitated for every dataset individually (Fig. 7E–G and Supplementary information 3). Genes had been filtered for these considerably variable between varieties (adjusted p < 0.05 and log fold change > 0.5) in a minimum of one dataset after which the modifications had been in contrast throughout environments. The highest correlation was evident between in vitro and mouse-derived datasets (r = 0.73, p < 0.05), with modifications between slender and stumpy-like varieties displaying barely decrease correlation between these information and cattle samples (cow vs. mouse: r = 0.59, p < 0.04; cow vs. in vitro: r = 0.68, p < 0.05).
Comparing slender-associated transcripts, 246 genes had been upregulated in slender varieties throughout all datasets (Supplementary information 3). The prime frequent markers consisted of genes linked to cell proliferation (e.g. histone H2B, chromosome passenger advanced 1 and cytoskeleton elements), quite a few RNA-binding proteins (e.g. RBP10, ZC3H38, ZC3H39, ZC3H31) and a non-coding RNA Tb1.NT.27. Just 13 genes had been uniquely upregulated in slender-like varieties within the cattle information solely in comparison with each in vitro and mouse information (Fig. 7E, F). Of these, 5 are additionally markers of cow cluster 3 described above: a 2-oxoglutarate dehydrogenase E1 part (Tb927.11.9980), aquaglyceroporin 1 (Tb927.6.1520), a calpain-like cysteine peptidase (Tv11.v5.1011) and two hypothetical proteins (Tb927.11.18710 and Tb927.7.790). The remaining eight genes encode 5 hypothetical proteins (see Supplementary information 3), a second putative calpain-cysteine peptidase (Tb927.11.1130), F-box protein FBP75 (Tb927.5.700) and a putative calcium motive p-type ATPase (Tb927.9.15460).
A standard stumpy-associated transcriptome was additionally clear (Fig. 7E–G), with probably the most conserved stumpy/stumpy-like markers together with mitochondrial-encoded cytochrome oxidase subunit 2 (COII), procyclin proteins (EP1, EP2, GPEET), PAD2, a number of copies of retrotransposon hotspot protein 4 (RHS4) and a number of other hypothetical proteins (see, Supplementary information 3). 422 genes confirmed increased transcript ranges in stumpy-like varieties in cattle, however not in stumpy varieties remoted from mice or in vitro. These included cAMP response protein 9 (CARP9; Tb927.8.4640), amino acid transporter 1 (AATP1; Tb927.8.7640), ESP9 (enriched in surface-labelled proteome protein 9; Tb927.9.11480), two putative metallopeptidase (FtsH33, Tb927.4.3300 and Tb927.11.3490), seven adenylyl cyclase (GRESAG 4.4) copies and members of the PAD array.
Given these host-specific variations, we tried to adapt cultured T. brucei strains to cattle serum in vitro and assess morphological parameters utilizing microscopy. Parasites couldn’t be tailored to twenty% cow serum as a substitute for 20% foetal calf serum (FCS) in HMI-9 medium. However, Antat 1.1 90:13 T. brucei could possibly be efficiently tailored to a combination of 10% cow serum and 10% FCS in HMI-9 after one week of tradition, at which level the parasites exhibited the identical progress charge as these cultured in 20% FCS (Supplementary Fig. 8A). At excessive density (>1.5 × 10⁶ parasites/ml), there have been no observable variations within the proportions of 1N1K parasites or in flagellum size (Supplementary Fig. 8B, C). PAD1 staining revealed no constructive parasites in 20% FCS, and seven out of 245 parasites had been PAD1-positive within the 10% FCS/10% cow serum situation throughout three replicates. Therefore, the addition of uninfected cattle serum alone to in vitro cultures of T. brucei doesn’t recapitulate the traits noticed in parasites remoted from cattle.
This web page was created programmatically, to learn the article in its authentic location you may go to the hyperlink bellow:
https://www.nature.com/articles/s41467-025-64750-y
and if you wish to take away this text from our web site please contact us