This web page was created programmatically, to learn the article in its authentic location you’ll be able to go to the hyperlink bellow:
https://www.nature.com/articles/s41540-025-00557-3
and if you wish to take away this text from our web site please contact us
Multi-omics information combines quantifying data of varied biomolecules and molecular processes that represent a cell or tissue. By contemplating these totally different, interacting processes, multi-omics information exhibit the potential to unravel extra insights into phenomena of curiosity over utilizing solely single-omics information. One such phenomenon that may notably profit from a multi-omics based mostly evaluation is most cancers, an space that’s not effectively understood and the place such information might provide significant insights.
Colorectal most cancers (CRC), the third commonest most cancers on the earth, is chargeable for ≈ 9% of all cancer-related deaths worldwide in 20221. To cut back the mortality charge, it’s crucial to grasp its molecular foundation to establish efficient therapy methods. A serious research carried out by the Consortium for Colorectal Cancer Subtyping utilised gene expression information to establish 4 consensus molecular subtypes (CMS) of CRC2. While the research resulted in a high-level characterisation of CRC subtypes, extending it to multi-omics solely produced partially comparable clusters3. The multi-omics-derived clusters outperformed CMS clusters in predicting affected person survival, which confirmed no prognostic worth for the cohorts of their research (TCGA COADREAD and CCLE)3. This prompted us to conduct a multi-omics evaluation of CRC within the context of the rarely-studied disease-specific survival (DSS).
However, dealing with multi-omics information comes with its challenges. It has been proven that utilizing multi-omics information in its entirety doesn’t have a lot profit for survival prediction over utilizing scientific or gene expression information, both resulting from interference from noise or excessive dimensionality of the information4,5. We exploit signatures recognized in prior research from every omics information to deal with these challenges. Our principal goal is to evaluate the potential of counting on multi-omics information to acquire related illness characterisation insights. In this context, relying solely on unsupervised approaches ensures limiting biases arising from taking a task-specific perspective to the issue. To this finish, we carry out unsupervised clustering over these multi-omics signatures and establish a 0-event group with considerably totally different survival behaviour (Fig. 1). In addition to scientific characterisation of the obtained teams, an in-depth evaluation of this considerably totally different survival group via evaluation of variance (ANOVA) and gene set enrichment evaluation (GSEA) highlights the necessary contributing options and the related pathways, offering extra insights into the clusters generated by multi-omics information. We prolong multi-omics information to incorporate complete slide photos and observe comparable outcomes.
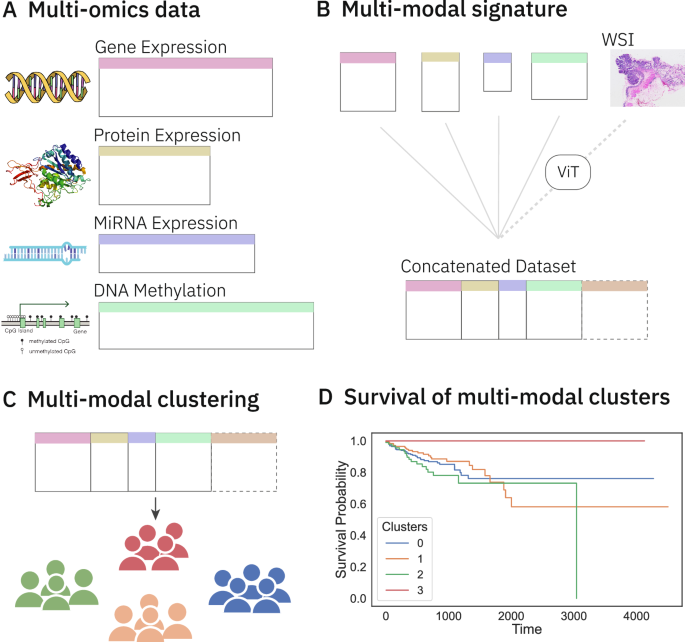
A Multi-omics information pertaining to colorectal most cancers (CRC) is collected from TCGA. B Signatures for every omics kind are retrieved from the literature. These signatures are used to subset every omics dataset, that are then concatenated to kind a multi-omics dataset. Whole slide picture (WSI) embeddings from a Vision Transformer (ViT) are concatenated to this multi-omics dataset to kind a multi-modal dataset. C Unsupervised clustering on these datasets teams sufferers into distinct clusters. D The clusters are analysed for distinctive survival patterns via survival evaluation strategies comparable to Kaplan-Meier curves and significance testing by Peto-weighted pairwise log-rank take a look at. Icons from Wikimedia Commons: DNA helix by Leyo (public area); miRNA by DBCLS (CC BY 4.0); protein by Emw (CC BY-SA 3.0); WSI by Ed Uthman, MD (CC BY-SA 2.0); DNA methylation by Mariuswalter (CC BY-SA 4.0).
Our objective is to point out that multi-omics information have advantages in stratifying sufferers to acquire related illness characterisation insights over single omics. To this finish, we work instantly with recognized CRC signatures from every omics information and consider this on DSS. Despite utilizing DSS as a criterion for analysis, we present how an unbiased and unsupervised method to multi-modal information evaluation naturally results in insights not obvious when utilizing single modalities.
Selecting extremely related options versus compressing all options right into a lower-dimensional area has its benefits. First, any inferences made on account of those options usually are not depending on the standard of transformations, as within the case of compression. Second, retaining the unique values makes deciphering the outcomes simpler. Third, the discount to a smaller, related subset minimises the chance of introducing spurious relationships. And 4, the influence of irrelevant and redundant options on downstream duties is diminished. For gene expression, we use 40 genes recognized by ref. 6 as being considerably related to CRC and the outlined CMS. For DNA methylation, we choose all of the probes mapping to differentially methylated genes related to CRC prognosis as present in refs. 7,8, along with 16 CpG websites recognized by ref. 9, 26 markers that may distinguish between CIMP-Negative and CIMP-Low tumours by ref. 10 and 5 markers which are differentially expressed in comparison with regular adjoining tissue by ref. 11, amounting to a complete of 82 probes in widespread with the TCGA COADREAD dataset. A set of miRNA signatures discovered to be considerably related to CRC development and metastasis12,13,14, together with a miRNA signature that may discriminate early stage CRC15,16,17, and a novel tumour suppressor miRNA18, make up the 30 miRNAs chosen for the research. Lastly, two main proteomics research19,20 and a research relating proteomics to prognosis21 led to the discovering of 11 proteins differentially expressed in CRC for our research.
To spotlight the advantage of combining totally different omics information in affected person stratification, we use CMS2 because the baseline for our analyses. Grouping sufferers by CMS ends in 4 totally different clusters, one for every subtype. In this experiment, we evaluate the distribution of DSS in baseline CMS clusters towards clusters created by similarity in (i) a unimodal setting: a subset of gene signatures related to CMS, (ii) a multi-omics setting: comprising (i) and choose markers from different omics information comparable to DNA methylation, miRNA expression and protein expression, and (iii) a multi-modal setting: comprising (ii) and complete slide photos. The optimum variety of clusters for every dataset is decided utilizing the elbow methodology, and is discovered to be Okay = 4 for all datasets, in keeping with the CMS groupings. The outcomes of the elbow methodology are proven in Supplementary Fig. S3, and Supplementary Table S2 summarises the common silhouette rating for the variety of clusters thought of to confirm the standard of the clusters. Silhouette rating analyses and PCA visualisations for the chosen optimum variety of clusters are illustrated in Supplementary Fig. S4. We moreover research the steadiness of the clusters generated by (ii) and (iii) by repeating the experiment 5 instances, every time with a distinct seed. The outcomes of this evaluation are mentioned in Supplementary Note S2.1. From the unimodal datasets, we choose gene expression for comparability as it’s the most generally used omics information. For reference, outcomes of the opposite unimodal datasets, particularly, DNA methylation, protein expression, miRNA expression, and complete slide photos are mentioned in Supplementary Note S3.
We look at survival features of the totally different clusters with the assistance of Kaplan-Meier plots (Fig. 2) and compute their pairwise significance utilizing the Peto-weighted pairwise log-rank take a look at (Table 1). We see that each multi-modal and multi-omics datasets establish a cluster of sufferers that have no occasion in any respect (Fig. 2a, b, respectively). The all-surviving cluster 3 recognized by multi-omics information has a considerably higher survival charge than cluster 0 (p = 0.01), cluster 1 (p = 0.01), and cluster 2 (p < 0.005). Similarly, the multi-modal dataset isolates cluster 0 because the all-surviving group, and this cluster has a considerably higher survival charge in comparison with cluster 1 (p < 0.005), cluster 2 (p = 0.01), and cluster 3 (p < 0.005). The addition of complete slide photos to multi-omics improves the general significance of variations between clusters by barely altering cluster composition.
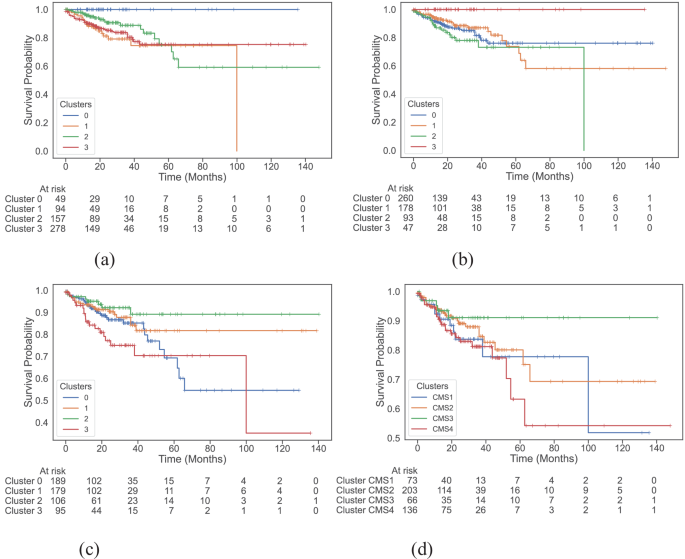
Kaplan-Meier plots of disease-specific survival (DSS) throughout clusters discovered by a multi-modal dataset, b multi-omics dataset, c colotype gene signatures, and d consensus molecular subtypes. The multi-modal and multi-omics datasets are in a position to establish an all-surviving cluster of sufferers with excessive significance.
Further observations of those Kaplan-Meier curves (Fig. 2) present distinguishable survival tendencies between clusters with time. For occasion, within the clusters generated by the multi-omics dataset (Fig. 2b), roughly after 15 months, the survival likelihood of sufferers in cluster 2 is decrease than sufferers in different clusters, and the survival likelihood of sufferers past 10-years on this cluster is 0, suggesting worse 10-year survival in comparison with the remaining. The same pattern is displayed by clusters from the multi-modal dataset, the place sufferers in cluster 1 have a survival likelihood of 0 past 10 years.
We then look into the options contributing to the clusters by performing an ANOVA take a look at between all cluster pairs. A binary heatmap of the highest 10 discriminating options between cluster pairs for the multi-omics information (Supplementary Fig. S5) and multi-modal information (Supplementary Fig. S6) exhibits genes and DNA methylation probes as main contributors. For insights into the all-surviving cluster, we carry out a gene set enrichment evaluation towards KEGG (2021)22,23 and MSigDB Hallmark (2020)24,25 gene units. In the case of multi-omics, we choose the union of all options which are considerably totally different (corrected p-value < 0.001) between clusters 0-3, 1-3, and 2-3, as cluster 3 is the all-surviving cluster. The discriminating options of cluster 3 are considerably enriched (adjusted p-value < 0.05) in 4 MSigDB Hallmark gene units – (i) Unfolded Protein Response (adjusted p-value 0.0405), (ii) UV Response Downregulation (adjusted p-value 0.0405), (iii) Epithelial Mesenchymal Transition (EMT) (adjusted p-value 0.0418) and, (iv) G2-M Checkpoint (adjusted p-value 0.0418). The unfolded protein response signalling pathways can swap cell survival to cell demise beneath endoplasmic reticulum stress, and their affiliation with colorectal most cancers is basically understudied26. Similarly, the function of genes downregulated resulting from UV radiation, comparable to receptor tyrosine kinases27 are additionally understudied, and findings from this multi-omics based mostly clustering warrant a deeper investigation into the function of those two processes on colorectal most cancers prognosis and development. The EMT course of has been related to most cancers development28, and research have investigated therapies towards it29. This course of might doubtlessly be comparatively downregulated in sufferers belonging to cluster 3. The G2-M checkpoint prevents cells from coming into mitosis when DNA is broken, and a steady G2 arrest helps to guard the genome and suppress tumourigenesis30. We repeat the identical experiment with the multi-modal clusters and discover the identical outcomes however with greater significance – (i) Unfolded Protein Response (adjusted p-value 0.0399), (ii) UV Response Downregulation (adjusted p-value 0.0399), (iii) Epithelial Mesenchymal Transition (EMT) (adjusted p-value 0.0411) and, (iv) G2-M Checkpoint (adjusted p-value 0.0411).
Given our findings, we carry out a couple of sanity checks on our information to validate our outcomes. First, we repeat the multi-omics clustering experiments with solely sufferers who’ve all modalities recorded. This case additionally elicits an all-surviving affected person group, thus ruling out any imputation artefacts (Supplementary Fig. S7). Additionally, we evaluate the highest 10 discriminating options on this case to those obtained from imputed multi-omics information, and discover important overlap (Supplementary Note S2.3). Second, because the clustering outcomes of multi-omics and multi-modal information are comparable, we decide if there are any redundancies between them by computing the correlation between pattern distances within the omics and picture areas (Supplementary Note S2.4) and discover no correlations (pearsonr statistic = 0.093, p worth=7.51e−127). Third, we carry out a clinico-pathological characterisation of the clusters obtained from each multi-omics and multi-modal information (Supplementary Figs. S10–S20) to not solely present that confounding components comparable to age and gender haven’t any impact on the clusters but additionally to spotlight the utility of multi-omics signatures in stratifying key scientific variables (Supplementary Note S2.5). Fourth, we verify for any data loss resulting from representing complete slide photos by a simplified imply of all patches. To this finish, we implement a bag-of-patches method based mostly on bag-of-visual-words31,32,33 utilizing each normalised counts and time period frequency-inverse doc frequency metrics to create two variations of representations that seize native data. On repeating the multi-modal evaluation utilizing each representations, we discover no variations within the clustering outcomes apart from a rise in correlation of pattern distances from 0.093 to roughly 0.17 between the omics and picture areas (Supplementary Note S2.6). Fifth, and eventually, we validate our findings on a second dataset – CPTAC34. As there’s a mismatch of obtainable modalities between TCGA and CPTAC, we use a proxy to assign cluster labels to CPTAC sufferers. A Kaplan-Meier survival evaluation with respect to overall-survival (OS) reveals similarities to TCGA-based multi-omics clusters – an all-surviving cluster (albeit with only one affected person) and a cluster having survival likelihood 0 after roughly 45 months (Supplementary Fig. S25). This proxy research is mentioned in Supplementary Note S2.7.
Comparing our findings to the baseline CMS (Fig. second), which aren’t considerably totally different from each other when it comes to DSS (Table 1), we see an enchancment. The prognostic energy of CMS is restricted to CMS4, which is related to worse general survival2. However, sufferers may exhibit blended CMS resulting from intra-tumour heterogeneity, which makes relying solely on the gene-expression-based CMS courses inadequate for prognosis35. To additional exhibit the advantages of utilizing a multi-omics-based method, we plot the distribution of CMS within the clusters recognized by multi-omics and multi-modal information (Supplementary Fig. S15). The poor 10-year survival cluster has a majority of CMS4 sufferers adopted by CMS1 sufferers, which is consistent with earlier findings2,36. However, we see that the all-surviving cluster is primarily a mixture of CMS1 and CMS3 clusters with no CMS2 sufferers, regardless of it being related to the very best general survival2. We consider multi-omics signatures are in a position to seize heterogeneity within the information, particularly given the impact of the complicated interaction between BRAF mutation, KRAS mutation, microsatellite instability (MSI), and CpG-island methylator phenotype (CIMP) on survival37,38. For occasion, the separation of CMS1 sufferers into the poor and all-surviving clusters means that multi-omics signatures appear to keep in mind this complicated interaction between excessive MSI and different components. Moreover, by instantly working with gene signatures as an alternative of CMS, we already see important variations within the survival charge between some clusters as seen of their Kaplan-Meier plot (Fig. 2c, Table 1). Thus, given the opportunity of sufferers having heterogeneous CMS characterisations and with no technique to at present handle this, the multi-view offered by multi-omics information might assist seize this intra-tumour heterogeneity, main to raised survival characterisation.
The advantage of our findings from multi-omics and multi-modal datasets lies within the potential development of personalised drugs analysis. The identification of a considerably totally different cluster distinctive to sufferers who expertise no occasions with each multi-modal and multi-omics information (Table 1) means that the present therapy technique is doubtlessly engaged on this group of sufferers. Even if these sufferers weren’t topic to any therapy, it could imply that their basic prognosis is optimistic, and additional analysis in therapies for CRC ought to be directed in direction of different teams, notably these clusters with a seemingly poor 10-year survival charge. This highlights the utility of incorporating data from a number of sources in affected person stratification. Information from a single supply might be biased, particularly if it isn’t but understood how the totally different components that make up our complicated system work together.
In conclusion, the inclusion of knowledge from different complicated organic programs in any evaluation is usually restricted by the variety of observations. But this does and shouldn’t stop us from exploring their mixed utility in getting one step nearer to understanding organic phenomena. Although we take the classical method of performing characteristic choice first by deciding on markers which were discovered to be related to CRC in prior works, a number of strategies exist that may carry out characteristic choice mechanically and study non-linear relationships between the options that might be helpful for affected person stratification. Future research might rigorously discover these strategies. Another avenue for additional evaluation is within the mixture of signatures that one selects from totally different omics information. It is believable that totally different mixtures of options might produce higher or worse stratification, and a research on this sensitivity to characteristic relevance would supply a dependable guideline to characteristic choice for affected person stratification.
This web page was created programmatically, to learn the article in its authentic location you’ll be able to go to the hyperlink bellow:
https://www.nature.com/articles/s41540-025-00557-3
and if you wish to take away this text from our web site please contact us



